Weekly Tech+Bio Highlights #23
ALSO: New EGFR-binding Proteins, Designing Novel IBD Treatment Using Gen-AI, Toward AI-Driven Digital Organism...
Hi! This is BiopharmaTrend’s weekly newsletter, ‘Where Tech Meets Bio,’ where we talk about technologies, breakthroughs, and great companies moving the biopharma and medtech industries forward.
If you've received it, then you either subscribed or someone forwarded it to you. If that is the case, subscribe by clicking this button:
Let’s get to this week’s topics!
Brief Insights
🔬 Eli Lilly plans large-scale trials in 2025 to explore its obesity drugs for addiction and cognitive disorders, signaling potential expansion of GLP-1 agonist applications to brain health, alongside ongoing investigations into cardiovascular and metabolic diseases.
🔬 Insilico Medicine nominates its 21st pre-clinical candidate and a BBB-penetrant NLRP3 inhibitor, to target inflammation-related and CNS diseases such as Alzheimer’s and epilepsy, leveraging its AI-driven Pharma.AI platform for advanced drug design.
🔬 Cradle wins Adaptyv Bio’s protein design challenge, creating an EGFR-binding protein with 8x improved affinity (1.21 nM KD) over Merck’s Cetuximab, showcasing its AI-driven platform’s precision in optimizing protein frameworks for cancer therapy targets.
🔬 BenevolentAI restructures to refocus on its techbio origins, shifting from late-stage biotech ambitions to early-stage drug development partnerships, causing layoffs while extending its cash runway into 2027.
💰 Pleno secures $25M to support the 2025 launch of its Raptor benchtop multiomics instrument, capable of analyzing DNA, RNA, and proteins, while appointing former Illumina executive Vik Vaz as CEO to lead its commercialization efforts.
💰 Citryll raises €85M in Series B funding to advance CIT-013, a monoclonal antibody targeting Neutrophil Extracellular Traps (NETs), into Phase 2a trials for rheumatoid arthritis and hidradenitis suppurativa by 2025.
🚀 Chroma Medicine and Nvelop Therapeutics merge to form nChroma Bio, securing $75M to advance CRMA-1001, a liver-targeted epigenetic editor for chronic hepatitis B and D, and develop next-gen genetic medicines with non-viral delivery technologies.
🔬 Owkin releases FedPyDESeq2, an open-source federated learning tool for secure differential expression analysis in RNA-seq studies, enabling collaborative research across siloed datasets while preserving data privacy and regulatory compliance.
🔬 Relation Therapeutics partners with GSK in a $45M upfront deal, with up to $200M per target in milestones, to develop treatments for fibrotic diseases and osteoarthritis using its AI-driven Lab-in-the-Loop platform to uncover patient-centric therapeutic targets.
🔬 Relay Therapeutics reports updated interim data for RLY-2608, its allosteric, mutant-selective PI3Kα inhibitor, showing improved PFS (11.4 months) and ORR (67%) in HR+/HER2- metastatic breast cancer, with plans for pivotal trials in 2025.
💰 Elevation Oncology partners with Synaffix in a $368M licensing deal to advance EO-1022, a HER3-targeting ADC for solid tumors, leveraging Synaffix’s GlycoConnect and toxSYN technologies for improved efficacy and safety, with an IND filing expected by 2026.
🚀 Xaira Therapeutics expands leadership with Dr. Paulo Fontoura as CMO and co-founder Dr. Hetu Kamisetty as CTO, while relocating to new headquarters in South San Francisco, signaling growth in AI-driven drug discovery and development.
🚀 Tasca Therapeutics launches with $52M in Series A to advance its lead candidate, CP-383, targeting auto-palmitoylation in difficult-to-treat cancers, into Phase 1/2 clinical trials by 2025.
🔬 Evaxion Biotech demonstrates preclinical proof-of-concept for AI-driven cancer vaccines targeting endogenous retrovirus (ERV) antigens, showing T-cell activation and tumor growth reduction, with plans to identify a lead candidate by late 2025.
🔬 Candid Therapeutics partners with EpimAb Biotherapeutics in a deal potentially exceeding $1B to develop novel bispecific T-cell engagers for autoimmune diseases, leveraging EpimAb’s FIT-Ig platform and Candid’s expertise in autoreactive B-cell targeting.
💰 BeiGene secures global rights to SYH2039, a MAT2A inhibitor targeting MTAP-deleted cancers, in a $150M licensing deal with CSPC Zhongqi, enhancing its solid tumor pipeline with plans for combination therapy alongside its PRMT5 inhibitor BGB-58067.
🔬 Researchers at Chung-Ang University identify the tRNA fragment 5′-tRH-GlyGCC as a key player in cancer progression, showing potential as a biomarker for early detection and a therapeutic target, with antisense oligonucleotides demonstrating tumor regression in mouse models.
🔬 Dot Compliance launches the third generation of its AI-powered eQMS, featuring enhanced insights, personalized quality guidance, and proactive risk detection to streamline compliance and quality management in life sciences.
🔬 The UK’s Defence and Security Accelerator (DASA) launches a £1M competition to boost microbial forensic capabilities, seeking innovative technologies for biosecurity aligned with the UK’s 2023 Biological Security Strategy; submissions close February 18, 2025.
This newsletter reaches over 7.4K industry leaders from organizations like NVIDIA, Microsoft, Novo Nordisk, Pfizer, Novartis, FDA, the US Department of State, Illumina Ventures, and hundreds more.
Interested in sponsoring?
Contact us at info@biopharmatrend.com
New EGFR-binding Proteins
Cradle’s platform secured first place in Adaptyv Bio's protein design competition by engineering an Epidermal growth factor receptor (EGFR)-binding protein with a binding affinity of 1.21 nM, outperforming Merck’s Cetuximab (9.94 nM) by 8-fold. The challenge attracted 130 teams and 1,131 designs, with only two surpassing Cetuximab in binding affinity.
Adaptyv Bio’s challenge tasked participants with designing novel binders for EGFR, a key target in cancer therapies for conditions like non-small cell lung cancer and colorectal cancer. The goal was to push the limits of protein engineering with designs distinct from known antibodies, including Cetuximab.
Of the 1,131 submitted designs, 400 underwent lab testing via Surface Plasmon Resonance (SPR), a technique used to measure molecular interactions in real-time. While many teams used tools like RFdiffusion or BindCraft, Cradle relied on its proprietary AI platform. Their design reportedly outperformed Cetuximab and led the field by a wide margin.
Cradle optimized the scFv-format Cetuximab rather than starting from scratch. By protecting the antibody’s CDR (Complementarity-Determining Regions)—critical for binding—from modifications, their platform focused on refining the surrounding framework regions, enhancing stability and binding strength.
Using Cradle’s AI-driven automation, framework regions were selected based on structural analysis. Mutations were proposed and scored by the platform’s models, and designs were finalized and submitted overnight without manual input; this method diverged from traditional CDR-focused approaches.
Stef van Grieken discussed how Cradle’s AI models uncover non-intuitive mutations and enable faster optimization cycles in our spring interview.
Cradle’s final design achieved a KD of 1.21 nM, outperforming Cetuximab (9.94 nM) and the next best competitor (5.18 nM). In-vitro performance was validated through SPR testing, despite initially ranking lower (300–600) in in-silico scoring due to competition mechanics favoring shorter sequences.
Improved binding affinity is an important step in drug discovery, but advancing to a therapeutic candidate requires addressing stability, manufacturability, and immunogenicity—factors that ensure the molecule’s clinical viability, scalability, and safety.
While the result is promising, further iterative design and testing are needed to really progress toward clinical applications.
Designing Novel IBD Treatment Using Generative AI
Concurrently with nominating its 21st preclinical candidate, Insilico Medicine has advanced ISM-5411, a potential new treatment for inflammatory bowel disease (IBD).
Inflammatory Bowel Disease (IBD), a chronic and debilitating autoimmune disorder that includes ulcerative colitis and Crohn’s disease, impacts millions of lives globally. Characterized by chronic gut inflammation, intestinal barrier dysfunction, and increased risk of colitis-associated cancer, IBD remains an unmet clinical challenge. Current treatments, largely focused on anti-inflammatory drugs, offer symptomatic relief but fail to address epithelial barrier damage—a core issue in IBD progression. As a result, patients face the threat of disease recurrence, prolonged dependence on medications, and increased risk of colorectal cancer.
A shift in the therapeutic paradigm is now emerging. The focus is turning toward drugs that actively repair the intestinal barrier while simultaneously reducing inflammation. One promising strategy lies in modulating the Hypoxia-Inducible Factor (HIF) pathway by inhibiting prolyl hydroxylase domain (PHD) enzymes. While this approach has shown potential, prior attempts to develop systemic PHD inhibitors for IBD have failed due to significant off-target effects, notably cardiovascular and tumorigenic risks.
But the story of ISM-5411 (also known in earlier studies as ISM012-042) is different. It revolves around innovation powered by artificial intelligence (AI), a generative chemistry platform, and a risky but seemingly rewarding approach to drug development, involving a high stakes bet on novel targets and design workflows. Developed using Insilico Medicine's AI-driven Pharma.AI platform and its Chemistry42 engine, ISM-5411 might be one of the vivid examples redefining how small molecules are discovered and developed for complex diseases like IBD.
The Role of PHD Inhibitors in IBD Treatment
Prolyl hydroxylases (PHDs) regulate the stability of hypoxia-inducible factors (HIFs), a family of transcription factors that control genes essential for epithelial integrity, tight junction formation, and immune regulation. The HIF-PHD axis plays a critical role in intestinal homeostasis, with PHD inhibition stabilizing HIF proteins to promote barrier repair and immune balance.
PHDs, however, present a “double-edged sword.” While their inhibition supports epithelial barrier repair, systemic inhibition activates HIFs in tissues beyond the gut, leading to increased erythropoiesis and vascular endothelial growth factor (VEGF) production, raising cardiovascular risks. This challenge was exemplified by the clinical failure of GB004, a systemic Fe(II)-chelator-based PHD inhibitor, in Phase 2 trials for IBD. Systemic PHD inhibition became synonymous with safety risks, leaving the industry in need of a better approach.
The challenge was clear: develop a gut-restricted PHD inhibitor. A drug that stays in the gut, acts locally, and avoids the systemic side effects of its predecessors. This is where Insilico Medicine entered the scene.
The Birth of ISM-5411 – From Concept to Candidate
In 2021, Insilico Medicine launched a bold mission: design a novel PHD inhibitor that overcomes the limitations of systemic inhibitors.
The development path was unconventional but proved to be efficient. Insilico began by using its PandaOmics platform to identify PHD1 and PHD2 as viable drug targets for IBD. This choice was guided by data-driven insights into the genes’ role in barrier repair, immune suppression, and inflammation control.
The team then harnessed Chemistry42 to initiate structure-based drug design effort. Using known PHD2 complex structures (like those with Takeda-17, JPHM-2-16, and Molidustat) as templates, Insilico's scientists defined six critical pharmacophore points and seeded a privileged fragment (a benzonitrile group) as the chemical starting point.
This starting point was subjected to fragment-based growth using Chemistry42’s structure-based generative model. The process involved the iterative generation of thousands of potential PHD inhibitors, with AI-driven models filtering candidates for novelty, drug-likeness, and synthetic accessibility.
Hit generation and lead optimization followed, supported by SAR (structure-activity relationship) analysis and potency refinement using Alchemistry, which calculates binding free-energy estimates for PHD2-ligand complexes. Next, ADMET profiling predicted key drug properties like solubility, permeability, and systemic exposure risk. The result of this process was a single lead candidate: ISM-5411.
Within 12 months, Insilico had to synthesize and screen only 115 molecules to nominate a preclinical candidate, which seems remarkable in terms of effort/result ratio, which might indicate efficiency of its computational platform.
A Speedy Path To Clinical Trials
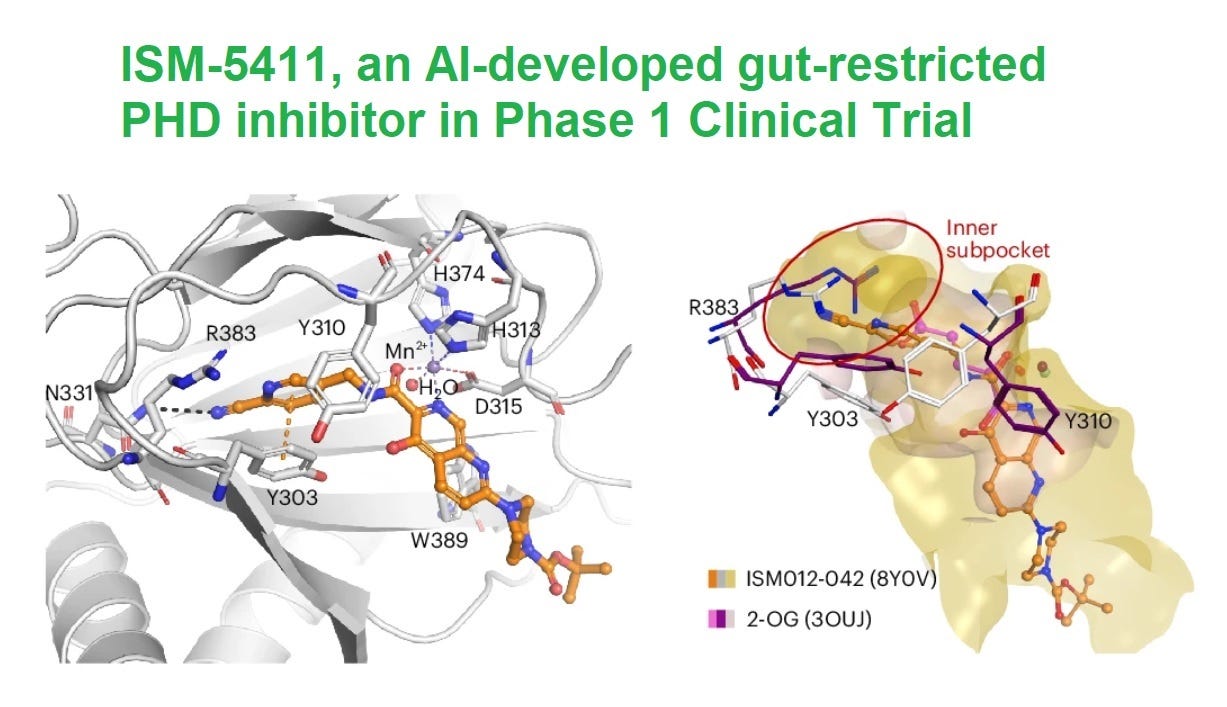
ISM012-042 stands out for its unique mechanism and pharmacological profile. The compound selectively inhibits PHD1 and PHD2 with IC50 values of 1.9 nM and 2.5 nM, respectively, while showing minimal activity on PHD3. This specificity addresses safety concerns, as PHD3 inhibition is linked to increased tumorigenic risk.
The most significant aspect of ISM012-042 is its gut-restricted pharmacokinetics. It remains in the colon, minimizing systemic exposure. Animal models confirmed a 67-fold higher concentration in the colon than in plasma, ensuring local activity where it is needed most. Pharmacokinetic analysis in rats revealed that nearly 85% of the compound was excreted in feces, reflecting low systemic absorption.
The compound's unique binding mechanism further distinguishes it from competitors. Unlike Fe(II) chelators like GB004, ISM012-042 binds PHD2 directly, inducing a conformational change that stabilizes HIF-1α, even in the presence of high iron concentrations. This feature eliminates the "iron dependence" of prior inhibitors.
To demonstrate efficacy, Insilico tested ISM012-042 in two experimental colitis models:
TNBS-induced colitis model (driven by chemical irritation)
Oxazolone-induced colitis model (mimicking human Th2-driven colitis)
Results were strong. ISM012-042 outperformed mesalamine, the current standard of care, in reducing disease activity index (DAI) scores and improving epithelial integrity. Colonic tissue samples revealed increased tight junction protein (ZO-1) retention and elevated expression of barrier-protective genes like TJP1, CD73, and TFF3. Cytokine profiling showed reductions in IL-6, IL-17, and TNF-α, while immune cell infiltration of neutrophils and pro-inflammatory T cells was significantly reduced. These effects were dose-dependent, supporting its potential for both preventative and therapeutic applications in IBD.
Armed with compelling preclinical data, Insilico Medicine submitted an Investigational New Drug (IND) application for ISM012-042. Approval was granted, leading to the initiation of a Phase 1 clinical trial (NCT06012578) in late 2023. This trial aims to assess the drug’s safety, pharmacokinetics, and pharmacodynamics in healthy human volunteers.
With its gut-restricted exposure, ISM012-042 is expected to avoid the cardiovascular and tumorigenic risks that derailed GB004. Early indicators suggest that the compound may finally achieve what other PHD inhibitors could not—a safe, effective, and gut-specific treatment for IBD.
Toward AI-Driven Digital Organism
In light of recent undertakings in foundation models, there’s a recent preprint by Le Song, Eran Segal, and Eric Xing where they propose a concept they call the AI-Driven Digital Organism (AIDO)—a computational system designed to simulate, predict, and program biological processes across scales, from molecular interactions to whole organisms.
The idea builds on multiscale foundation models but takes a broader view of biology's challenges. While physics and chemistry benefit from predictive frameworks—Newton’s laws for motion, the periodic table for elements—biology remains resistant to such systems, its complexity spanning molecules, cells, and entire organisms.
Despite recent advancements, most current foundation models in biology are designed and built for individual data modalities. They do not account for the multiscale nature of biology and the multimodal characteristics of biological data, and therefore, are not quite "foundational" for biology. Consequently, a truly foundational model which is intrinsically integrative, multiscale, and connected across diverse biological data, and is capable of addressing biological questions across different scales, is still missing. It is our view that a foundation model for biology—which can be a system of component FMs—needs to incorporate multiple types of data and biological constraints arising from different biological scales. Furthermore, such a system is more than just an agglomeration of modality-specific FMs, and must involve system-wide harmonization through nested or hierarchical representation propagation, utilization, fine-tuning, or continual pretraining. It should also have the ability to connect different FM modules from the system, and provide a foundation to address more complex prediction, simulation, and reprogramming tasks arising from molecules, cells, organisms, and beyond.
AIDO is presented as a step toward addressing this gap: a digital system designed to model life across scales, where experimentation can be simulated, tested, and refined safely and efficiently. The system is laid out in three modular stages:
Divide and Conquer: This phase involves constructing separate models for different biological scales, such as DNA, proteins, and cellular interactions. Existing models like AlphaFold and RNA-FM are cited as building blocks for this approach.
Connect the Dots: The authors outline how these modules could be integrated using techniques like graph-based computation and hierarchical encoding to simulate the interconnected nature of biological systems.
Piece It Together: Finally, the system is unified, allowing information to flow between levels and enabling optimization through feedback loops, aligning molecular-scale models with organismal-level predictions.
Although AIDO is still at a conceptual stage, the paper outlines its broad potential applications, such as simulating protein design and drug interactions in molecular engineering, predicting genetic or chemical effects on cellular systems, and linking genomic data to population-scale health outcomes for precision medicine.
The authors acknowledge significant challenges—the need for scalable computing infrastructure, harmonizing diverse datasets, and developing architectures capable of addressing the complexity of biological systems. Existing models for tasks like DNA or protein analysis remain highly specialized and limited in scope, falling short of the integrative, multiscale vision required to create a true "digital organism."
Read also:
11 Biopharma Trends to Watch in 2024